2024
Deep Learning - Making Medical Texts Meaningful

Clinical-Text-Classification
- Introduction
This project aims to classify medical text from medical transcriptions using advanced transformer models specifically adapted for clinical text. We utilize ClinicalBERT and Bio_ClinicalBERT, as well as a customized lightweight version of ClinicalBERT, to perform clinical text classification. The evaluation metrics include confusion matrix, accuracy, precision, recall, F1-score, Area Under the Curve (AUC), and Precision-Recall (PR) curves. - Data Review
The dataset used for this project is the Medical Transcriptions dataset available on Kaggle. This dataset contains various medical transcriptions categorized into different medical specialties.
Dataset link: Medical Transcriptions
The data set was examined, missing values in the text column were cleared and grouped according to medical characteristics. Word statistics were obtained. After examining the number of categories, categories with more than 50 numbers were determined.
A chart has been created showing the distribution of more than 50 categories.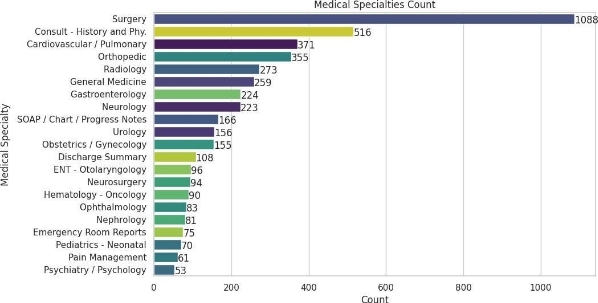
Two sample transcripts selected before data preprocessing are shown.Sample Transcription 1 HISTORY OF PRESENT ILLNESS: The patient is a 17-year-old female, who presents to the emergency room with foreign body and airway compromise and was taken to the operating room. She was intubated and fishbone. PAST MEDICAL HISTORY: Significant for diabetes, hypertension, asthma, cholecystectomy, and total hysterectomy and cataract. ALLERGIES: No known drug allergies. CURRENT MEDICATIONS: Percival, Humalin, Diprivan, Proventil, Unasyn, aspirin, regular Insulin, airway, Atrovent, Mom. FAMILY HISTORY: Significant for cancer, hospital for illicit drugs, alcohol, and tobacco. PHYSICAL EXAMINATION: Please see the hospital chart. LABORATORY DATA: Please see the hospital chart. RADIOGRAPHIC DATA: The patient was taken to the operating room by Dr. X who is covering the hospital and noted that she had airway compromise and a rather large fishbone could be seen in the esophagus. The airway was intubated and we will see if she should be observed to see if the airway would improve upon which could be the intubated. Dr. X had a large part of fishbone removed. The patient was treated with early antibiotics and ventilatory support and the toe of his arms dictation. She was resuscitated and taken to the operating room where it was felt that the airway was fixed and she was stable. She was doing well as of this dictation and is being prepared for discharge at this point. We will have Dr. X evaluate her before she leaves to make sure I do not have any problems with her going home. We feel she could be discharged today and will have her return to see him in a week. Sample Transcription 2 PREOPERATIVE DIAGNOSIS: Painful ingrown toenail, left big toe. POSTOPERATIVE DIAGNOSIS: Painful ingrown toenail, left big toe. OPERATION: Removal of an ingrown part of the left big toenail with excision of the nail matrix. DESCRIPTION OF PROCEDURE: After obtaining informed consent, the patient was taken to the minor OR room and intravenous sedation with morphine and versed was performed and the toe was blocked with 1% Xylocaine after having been prepped and draped in the usual fashion. The ingrown part of the toenail was freed from its bed and removed, then a flap of skin had been made in the area of the matrix supplying the particular part of the toenail. The matrix was excised down to the bone and then the skin flap was placed over it. Hemostasis had been achieved with a cautery. A tubular dressing was performed to provide a bulky dressing. The patient tolerated the procedure well. Estimated blood loss was negligible. The patient was sent back to Same Day Surgery for recovery. - Data Preprocessing
Text cleaning and lemmatization processes were carried out to better analyze and classify the text data of the project. The text data went through basic preprocessing steps and was prepared for classification. And finally, in this section, the metrics to be used to evaluate the classification models are defined.
Two sample transcripts selected after data preprocessing are shown.Sample Transcription 1 history of present illness the patient is a yearold female who present to the emergency room with foreign body and airway compromise and wa taken to the operating room we will have dr x evaluate her before she leaft to make sure i do not have any problem with her going home Sample Transcription 2 preoperative diagnosis painful ingrown toenail left big toe postoperative diagnosis painful ingrown toenail left big toe operation removal of an ingrown part of the left big toenail with excision of the nail matrix description of procedure after obtaining informed consent the patient wa taken to the minor or room and intravenous sedation with morphine and versed wa performed and the toe wa blocked with xylocaine after having been prepped and draped in the usual fashion estimated blood loss wa negligible - Models
We implemented three different models for clinical text classification:- ClinicalBERT
- Model link: ClinicalBERT
- Bio_ClinicalBERT
- Model link: Bio_ClinicalBERT
- Customized Lightweight ClinicalBERT
- We chose one of the three versions provided in the literature:
- Distil-ClinicalBERT
- Tiny-ClinicalBERT
- Clinical-MiniALBERT-312
For the purpose of this project, we used the Distil-ClinicalBERT version. - Methodology
The implementation follows the clinical text classification approach detailed in the Kaggle notebook by Rithesh Sreenivasan. The methodology involves:- Preprocessing the medical text data.
- Adapting the ClinicalBERT and Bio_ClinicalBERT models to classify the text.
- Training and evaluating the models using standard evaluation metrics.
- Comparing the performance of the three models.
Implementation reference: Clinical Text Classification - Evaluation Metrics
The performance of the models was evaluated using the following metrics:- Confusion Matrix
- Accuracy
- Precision
- Recall
- F1-Score
- AUC (Area Under the Curve)
- PR (Precision-Recall) Curves
- Results
Confusion Matrix
ClinicalBERT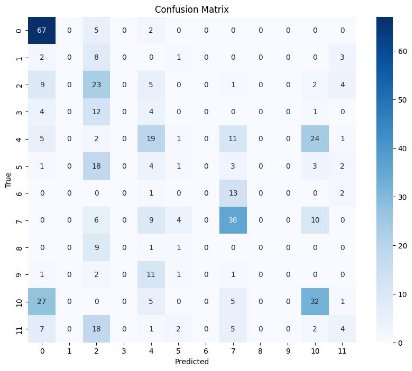
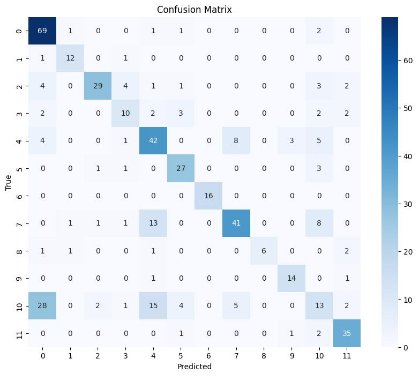
Distil-ClinicalBERT
AccuracyModel Accuracy ClinicalBERT 0.3913978494623656 Bio_ClinicalBERT 0.6752688172043011 Distil-ClinicalBERT 0.6580645161290323
Precision, Recall, F1-ScoreModel Precision Recall F1-Score ClinicalBERT 0.307 0.391 0.333 Bio_ClinicalBERT 0.666 0.675 0.657 Distil-ClinicalBERT 0.656 0.658 0.647
AUC and PR Curves
ClinicalBERT



Distil-ClinicalBERT

- Observations
- Best Overall Performance: Bio_ClinicalBERT demonstrated the best overall performance with the highest accuracy (0.675), precision (0.666), recall (0.675), and F1-score (0.657). This indicates that Bio_ClinicalBERT is highly effective in correctly classifying instances and minimizing false positives while accurately identifying true positives.
- Balanced Performance: Distil-ClinicalBERT also showed a strong performance, with accuracy (0.658), precision (0.656), recall (0.658), and F1-score (0.647). Although slightly lower than Bio_ClinicalBERT, it still maintained a balanced and robust performance.
- ClinicalBERT's Baseline: ClinicalBERT, with the lowest accuracy (0.391), precision (0.307), recall (0.391), and F1-score (0.333), highlighted the challenges in the dataset and served as a baseline model. The significantly lower metrics indicate that ClinicalBERT struggled with the complexity of the classification task.
- Lightweight Efficiency: Distil-ClinicalBERT's performance underscores the potential of lightweight models to achieve high accuracy and reliability while being resource-efficient. Despite its compact size, it maintained competitive performance metrics, making it a viable option for practical applications, especially in resource-constrained environments.
- Conclusion
Summary of the Project:- Best Performing Model: Bio_ClinicalBERT emerged as the best performing model with the highest accuracy, precision, recall, and F1-score. Its superior performance metrics indicate that it is highly effective in clinical text classification tasks.
- Worst Performing Model: ClinicalBERT, with the lowest metrics across accuracy, precision, recall, and F1-score, was the least effective model. This highlights the need for further fine- tuning or using more advanced models to handle the complexities of the dataset.
- Best Performance: Bio_ClinicalBERT's performance metrics (Accuracy: 0.675, Precision: 0.666, Recall: 0.675, F1-Score: 0.657) demonstrate its robustness and reliability in correctly classifying clinical texts.
- Worst Performance: ClinicalBERT's metrics (Accuracy: 0.391, Precision: 0.307, Recall: 0.391, F1-Score: 0.333) indicate significant room for improvement, particularly in handling the complexities of the dataset and reducing false positives.
This project illustrates the effectiveness of transformer models in clinical text classification, with Bio_ClinicalBERT showing the best overall performance. Distil-ClinicalBERT also proved to be a strong contender, offering a good balance between performance and efficiency. Future research could focus on further optimizing these models and exploring additional lightweight variants to enhance both accuracy and computational efficiency in clinical applications.